Project description
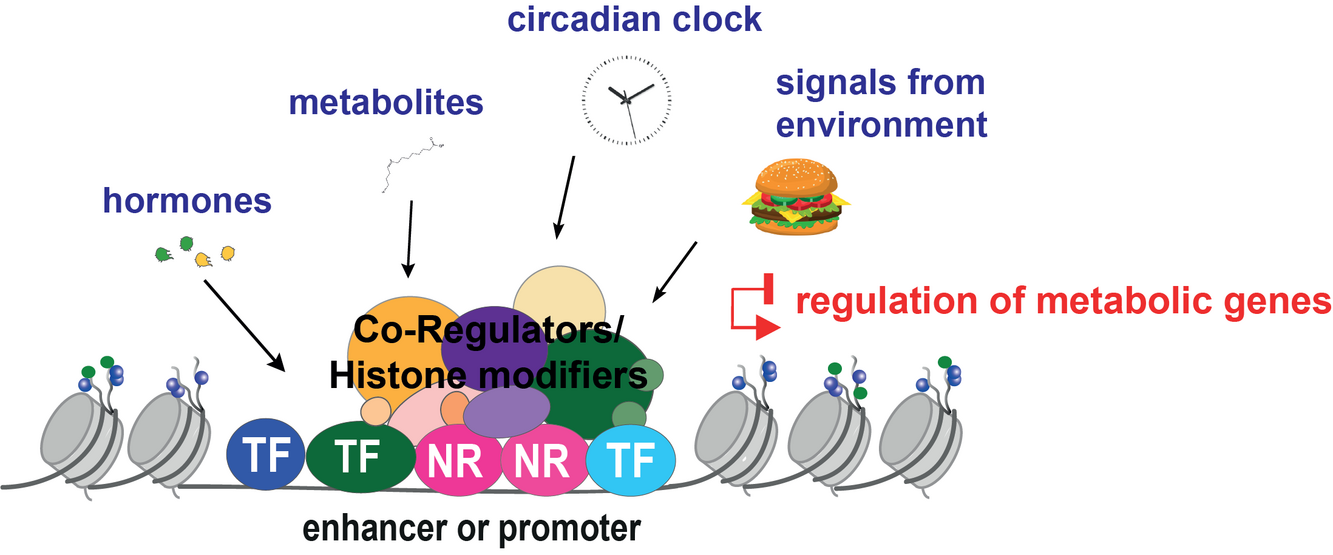
Excess of food intake is the leading cause of metabolic disease. Over-nutrition alters those metabolic programs required to maintain energy balance, by affecting nutrient and hormonal signaling. Crucial to this adaptation is the remodeling of target gene expression. The transcriptional control of metabolic homeostasis is primarily mediated by Nuclear Receptors (NR), a group of transcription factors that act as genomic switches, by directly responding to dietary lipids, hormones, and other intracellular signals. Once activated, NR bind to promoter and enhancer regions, recruit coregulator complexes, and regulate gene expression through mechanisms that largely depend on the epigenome editing. Hence, these transcriptional complexes function as metabolic sensors, capable of inducing histone modifications that dynamically reorganize the chromatin landscape depending on the environmental conditions. In our group we aim to elucidate the molecular mechanisms by which nutritional status influences NR genomic actions via the identification of specific genomic signatures and transcriptional profiles. The project will use interdisciplinary approaches including Next Generation Sequencing techniques (ChIP-seq, RNA-seq, ATACseq), mouse model of metabolic diseases, human genetics, and computational biology, in order to understand how the nutritional challenge and altered chromatin landscape affect the metabolic homeostasis. Upon completion, we will be able to address novel mechanisms of target gene regulation during the hepatic adaptation to over-nutrition, which could be ultimately exploited in the field of pharmacological research.
Relevant literature
- Escoter-Torres L, Greulich F, Quagliarini F, Wierer M, Uhlenhaut NH. (2020) Anti-inflammatory functions of the glucocorticoid receptor require DNA binding. Nucleic Acids Res. 48(15):8393-8407.
- Quagliarini F, Mir AA, Balazs K, Wierer M, Dyar KA, Jouffe C, Makris K, Hawe J, Heinig M, Filipp FV, Barish GD, Uhlenhaut NH. (2019) Cistromic Reprogramming of the Diurnal Glucocorticoid Hormone Response by High-Fat Diet. Mol Cell. 76(4):531-545. Research Highlight: Diet reprogrammes glucocorticoid rhythms. Starling, S. Nature Reviews Endocrinology 2020 V16, p.5.
- Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora E, Henderson B, Luu N-T, Portman J, Matchett K, Brice M, Marwick JA, Richard T, Efremova M, Vento-Tormo R, Carragher N O, Kendall TJ, Fallowfield JA, Harrison EM, Mole DJ, Wigmore SJ, Newsome PN, Weston CJ, Iredale JP, Tacke F, Pollard JW, Ponting CP, Marioni JC, Teichmann SA, Henderson NC. (2019) Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 575:512-8.
- Dyar KA, Hubert MJ, Mir AA, Ciciliot S, Lutter D, Greulich F, Quagliarini F, Kleinert M, Fischer K, Eichmann TO, Wright LE, Peña Paz MI, Casarin A, Pertegato V, Romanello V, Albiero M, Mazzucco S, Rizzuto R, Salviati L, Biolo G, Blaauw B, Schiaffino S, Uhlenhaut NH. Transcriptional programming of lipid and amino acid metabolism by the skeletal muscle circadian clock. (2018) PLoS Biol. 16(8):e2005886.
- Sirey TM, Roberts K, Haerty W, Bedoya-Reina O, Rogatti-Granados S, Tan JY, Li N, Heather LC, Carter RN, Cooper S, Finch AJ, Wills J, Morton NM, Marques AC, Ponting CP.( 2019) The long non-coding RNA Cerox1 is a post transcriptional regulator of mitochondrial complex I catalytic activity. Elife. May 2;8:e45051. doi: 10.7554/eLife.45051.
