Project description
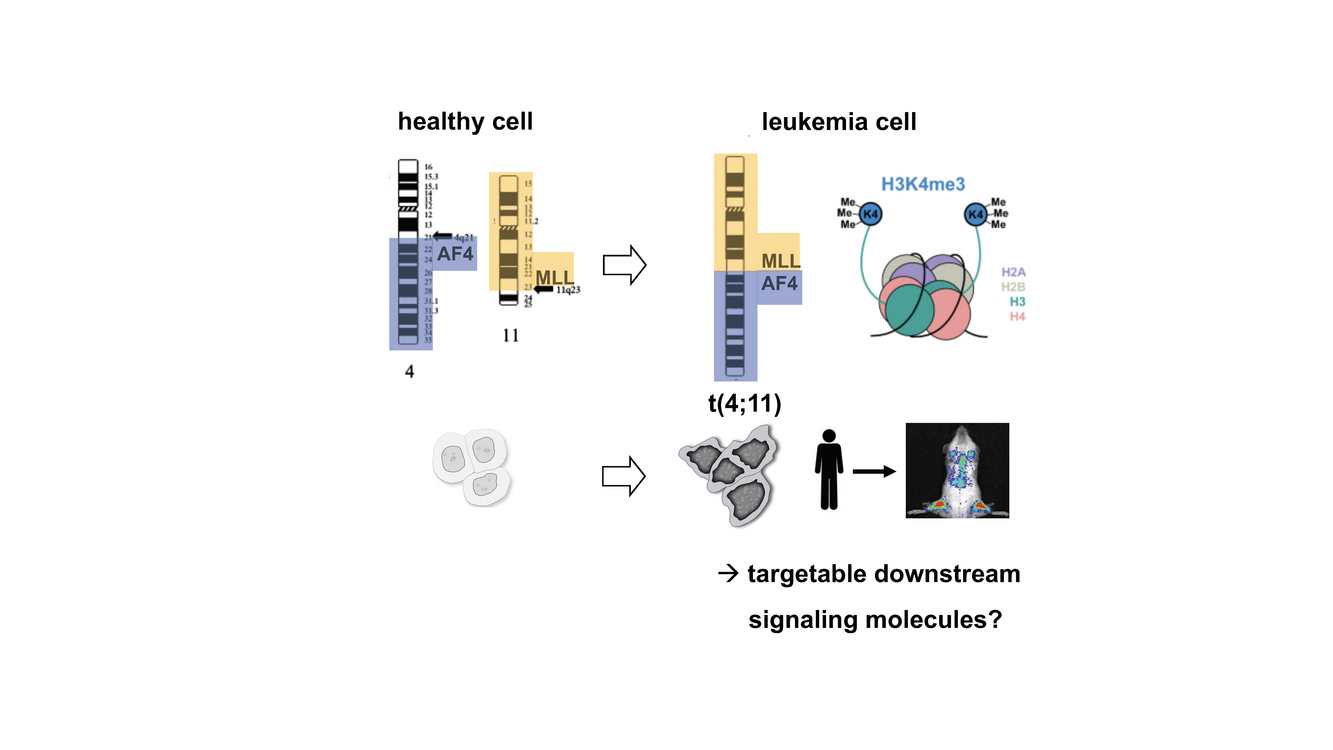
Translocation t(4;11) drives highly aggressive MLL-AF4 (KMT2A-AFF1) positive acute
leukemias, associated with poor prognosis. Due to the aberrant fusion of the two epigenetic
regulators, MLL and AF4, and recruitment of their associated complexes, MLL-AF4
positive leukaemia is characterized by multiple epigenetic changes which drive the
malignant disease. MLL is a positive regulator of gene expression by mediating e.g. H3K4
methylation. AF4 is the central scaffold protein of the super elongation complex (SEC),
which increases the catalytic rate of RNA polymerase II transcription. The transcriptional
changes caused by the MLL-AF4 driver mutation that result in various leukemic outcomes
are largely unknown. A better understanding of the mechanisms responsible for disease
progression is a prerequisite to developing innovative, targeted and efficient treatment to
improve the prognosis of patients.
Previously, the two PIs in Edinburgh and Munich have studied MLL-AF4 positive
leukaemia using complementary in vivo models. In syngeneic mice, we recently showed
that distinct microRNAs and their dysregulated targets are important downstream
mediators of MLL-AF4-driven leukemic growth and lineage choice (Malouf et al, Blood
2021). Using patient-derived xenograft models, we recently demonstrated that MLL-AF4
positive leukaemia remains dependent on the MLL-AF4 translocation, also when
established patient-derived xenograft (PDX) leukaemia grows in mice (Carlet et al., Nature
Comm. 2021).
We now plan to join forces in a search for downstream signaling molecules which are
upregulated by MLL-AF4 and relevant for leukaemia survival and growth. To study the
functional relevance of published and own candidate genes, we will use CRISPR/Cas9
knockout screens in existing MLL-AF4 positive PDX leukaemia models in vivo, followed
by single gene validation. Epigenetic changes induced by the genetic intervention will be
monitored using, e.g., ATAC sequencing or ChIP-Seq for H3K79 methylation, to
understand whether the leukemic phenotype could be reversed towards a physiologic
phenotype. As functionally relevant candidates represent putative therapeutic targets, we
will search compound inhibitors and perform preclinical treatment trials. At best, we can
translate the basic biological insights into clinical options for future use.
Overall, the project will lead to a better understanding of epigenetic changes in MLL-AF4-
positive leukemia and translate the knowledge into novel treatment options.
Relevant literature
1. Malouf C, Antunes ETB, O'Dwyer M, Jakobczyk H, Sahm F, Landua SL, Anderson RA, Soufi A, Halsey C and K.
Ottersbach (2021) “miR-130b and miR-128a are essential lineage-specific codrivers of t(4;11) MLL-AF4 acute
leukemia.” Blood. 2021 Nov 25;138(21):2066-2092
2. Symeonidou V, Jakobczyk H, Bashanfer S, Malouf C, Fotopoulou F, Kotecha RS, Anderson RA, Finch AJ and K.
Ottersbach (2021) “Defining the fetal origin of MLL-AF4 infant leukemia highlights specific fatty acid requirements.”
Cell Rep. 37(4):109900.
3. Carlet M, Völse K, Vergalli J, Becker M, Herold T, Arner A, Senft D, Jurinovic V, Liu WH, Gao Y, Dill V, Fehse B,
Baldus CD, Bastian L, Lenk L, Schewe DM, Bagnoli JW, Vick B, Schmid JP, Wilhelm A, Marschalek R, Jost PJ,
Miething C, Riecken K, Schmidt-Supprian M, Binder V and Jeremias I. (2021) „In vivo inducible reverse genetics in
patients' tumors to identify individual therapeutic targets.” Nat Commun. 2021 Sep 27;12(1):5655
4. Ebinger, S., E. Z. Ozdemir, C. Ziegenhain, S. Tiedt, C. Castro Alves, M. Grunert, M. Dworzak, C. Lutz, V. A. Turati,
T. Enver, H. P. Horny, K. Sotlar, S. Parekh, K. Spiekermann, W. Hiddemann, A. Schepers, B. Polzer, S. Kirsch, M.
Hoffmann, B. Knapp, J. Hasenauer, H. Pfeifer, R. Panzer-Grumayer, W. Enard, O. Gires and I. Jeremias (2016).
"Characterization of Rare, Dormant, and Therapy-Resistant Cells in Acute Lymphoblastic Leukemia." Cancer Cell
30(6): 849-862