Project description
Ribosomopathies are a group of congenital human diseases, which are caused by attenuated generation or activity of ribosomes. Patients’ cells possess reduced overall protein translation rates, ultimately resulting in complex pathologies.
We have developed several CRISPR based strategies enabling us to manipulate chromatin and gene expression at will. In a recently developed epigenome editing approach (TAPIR) we show that we can apply these tools to manipulate protein translation rates in mouse cell culture and living animals (Wiesbeck et al., in preparation). In this way, we have been able to prove that stem cell behavior during development is controlled by protein translation rates directly. Currently, we are investigating in detail, which chromatin mechanisms are causally involved in this process.
The suggested project will be conducted in collaboration with David Tollervey (Wellcome Centre for Cell Biology) and consists of two sub-projects. (1) We will apply TAPIR to a mouse model of ribosomopathy, recently established in the Tollervey lab (Robertson et al., 2021). (2) We will transfer, what we have learned from mouse cells and tissues to primary cells derived from ribosomopathy patients (and engineered iPSCs with patient mutations, available in the Tollervey lab). Since epigenome editing depends primarily on the targeting of dCas9 fusion proteins on gene regulatory elements, whose sequences are poorly conserved between species, the project will involve design of gRNA sequences and verification of their functionality. Other important aspects of the project are the adoption of in and ex vivo approaches and human iPSC culture. Taken together, this project will allow to comprehensively test, whether non-genetic CRISPR approaches are a suitable strategy to attenuate the effects of the ribosomopathy mutations.
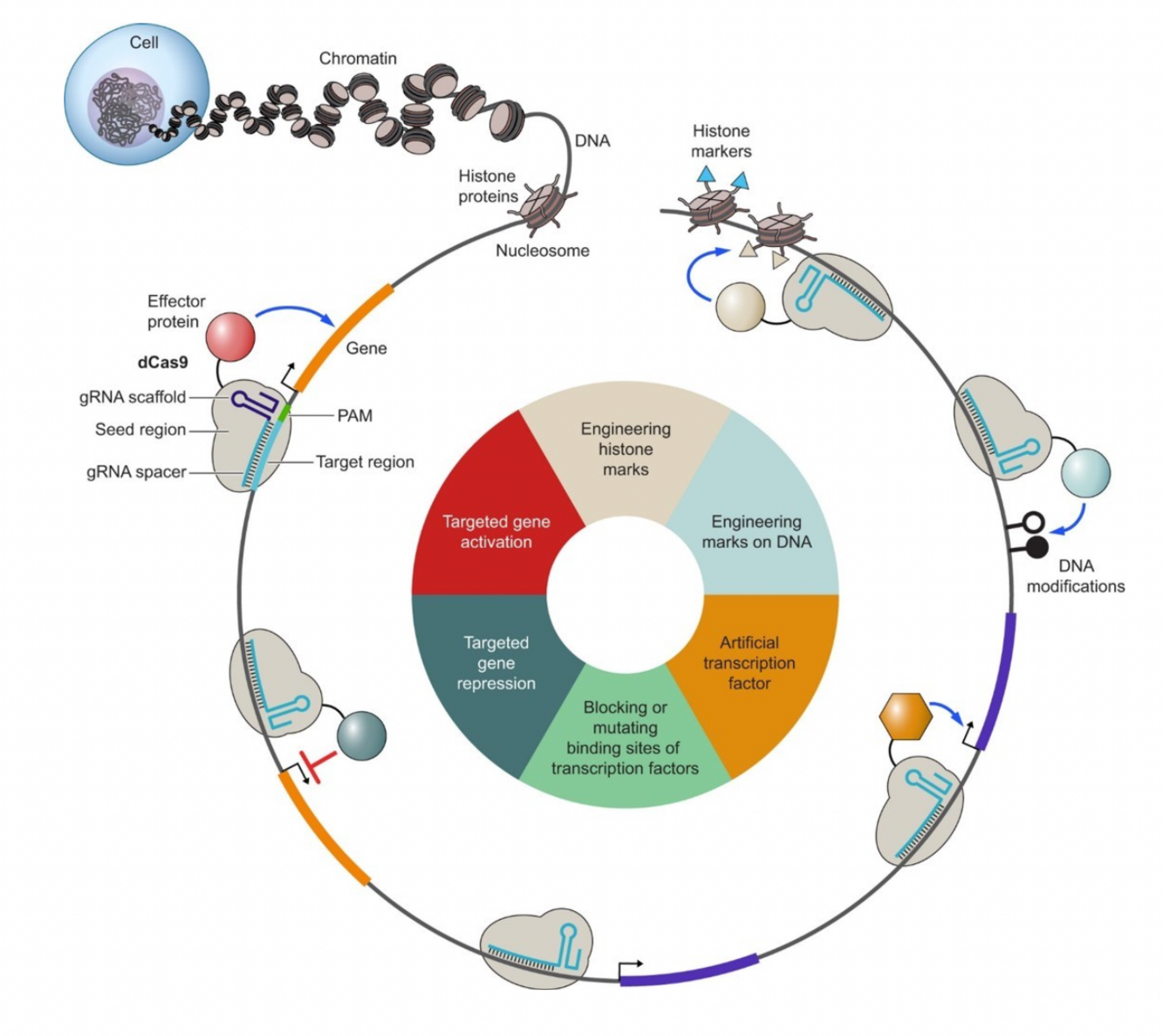
Figure legend: New molecular CRISPR tools to manipulate cellular states. The nucleus is the steering console of the cell. Molecular instructions for all possible cellular states are encoded in DNA. DNA is embedded in chromatin. CRISPR strategies include the activation and repression of critical genes, the interference with TF binding, the use of artificial transcription factors and the manipulation of chromatin features. Figure taken from Breunig et al., 2021.
Relevant literature
- Baumann, V., Wiesbeck, M., Breunig, C.T., Braun, J.M., Koferle, A., Ninkovic, J., Gotz, M., and Stricker, S.H. (2019). Targeted removal of epigenetic barriers during transcriptional reprogramming. Nature communications 10, 2119.
- Breunig, C.T., Koferle, A., Neuner, A.M., Wiesbeck, M.F., Baumann, V., and Stricker, S.H. (2021). CRISPR Tools for Physiology and Cell State Changes: Potential of Transcriptional Engineering and Epigenome Editing. Physiol Rev 101, 177-211.
- Clerget, G., Bourguignon-Igel, V., Marmier-Gourrier, N., Rolland, N., Wacheul, L., Manival, X., Charron, C., Kufel, J., Mereau, A., Senty-Segault, V., et al. (2020). Synergistic defects in pre-rRNA processing from mutations in the U3-specific protein Rrp9 and U3 snoRNA. Nucleic Acids Res 48, 3848-3868.
- Roberston, N. Shchepachev, V., Wright, D., Turowski, T.W., Spanos, C., Helwak, A., Zamoyska, R. and Tollervey, D. (2021) A disease-linked lncRNA mutation in RNase MRP inhibits ribosome synthesis. bioRxiv (https://biorxiv.org/cgi/content/short/2021.03.29.437572v1)
- Stricker, S.H., Koferle, A., and Beck, S. (2017). From profiles to function in epigenomics. Nat Rev Genet 18, 51-66.
