Project description
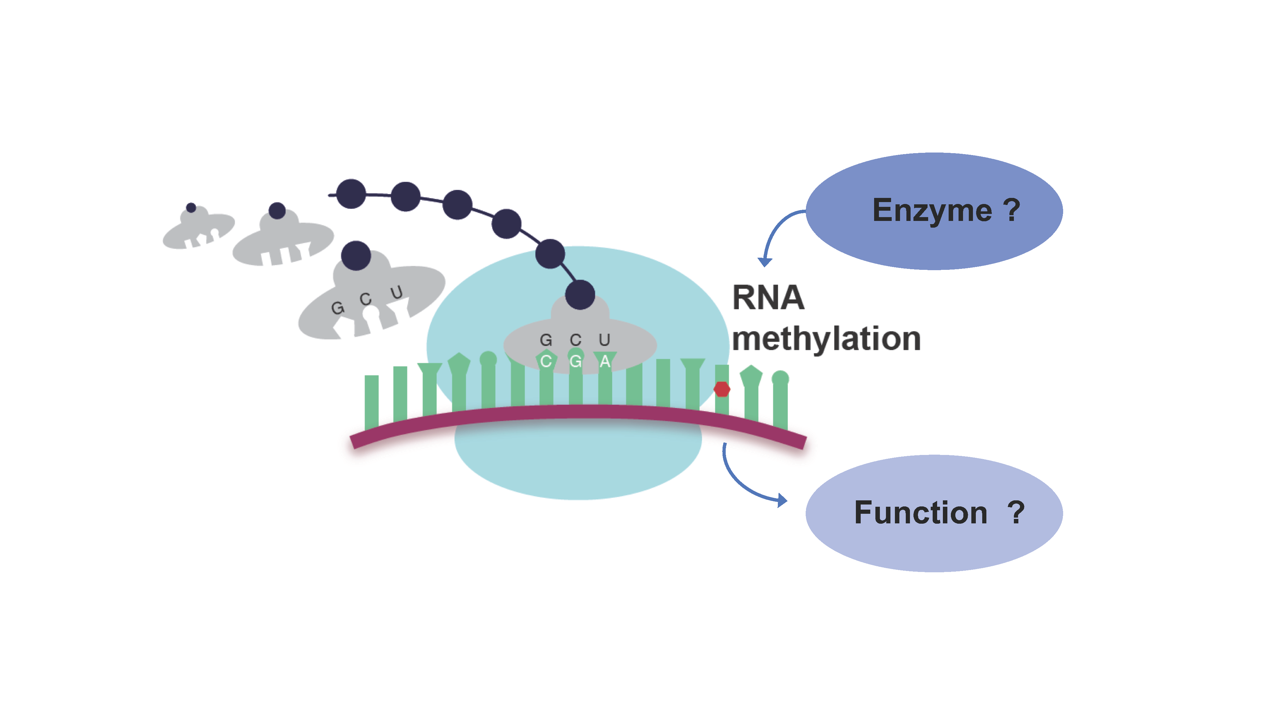
Recent breakthroughs have transformed our understanding of the epitranscriptome and allow us now to interrogate modifications of RNA at an unprecedented scale. It became clear that RNA modifications play essential roles in the regulation of splicing, nuclear export of RNA, RNA stability and translation. The epitranscriptome constitutes a new layer of (epi)genetic information and could change the way how we look at RNA, but also open new avenues to fight cancer. We recently identified two novel RNA methyltransferases and revealed their function in vitro and in vivo, creating new mouse models for human diseases (Ignatova et al., 2020a; Ignatova et al., 2020b).
The aim of the project will be i) to identify novel methyltransferases as well as ii) to determine the function of these enzyme(s) and iii) of the respective modifications in development and disease by applying a combination of in vitroand in vivo assays. For this project we will team up with the O’ Carroll lab. We will build on the expertise of the O’Carroll lab in cellular manipulations, mouse models and the role of RNA modifications in development and during hematopoiesis as well as on our expertise in the identification of novel chromatin and RNA modifying enzymes.
This project includes the possibility to be part of a dynamic and international team and to learn state of the art technologies both in Munich and Edinburgh (such novel NGS based mapping approaches and their bioinformatic analysis, as wells as Crispr/Cas9/Cas13 mediated manipulations, advanced imaging techniques, cellular assays, biochemical approaches and in vitro assays). It requires the motivation to develop and execute the next breakthrough ideas in an interdisciplinary and scientifically stimulating environment. We will employ diverse systems such as mES cells, mouse models and human cancer cells coupled with omics, advanced biochemistry and masspectrometry to gain insights in the mechanism of action of novel RNA methyltransferases. Since multiple studies have linked RNA modifications to cancer this project has the potential to discover novel druggable targets to treat cancers.
The over-arching long-term goal is to (i) understand mechanistically how RNA modifications regulate RNA, (ii) identify novel RNA modifying pathways in order to discover new therapy targets and (iii) move the fascinating epitranscriptomics field to a new level.
Relevan literature
- Ignatova, V.V., Kaiser, S., Sook Yuin, S., Bing, X., Stolz, P., Tan, Y.X., Xim Tan, Y., Leng Lee, C., Hoon Gay, F.P., Rico Lastres, P.,Gerlini, R., Rathkolb, B., Aguilar-Pimentel., A. Sanz-Moreno, A., Klein-Rodewald, T., Calzada-Wack, J.,Ibragimov, E., Valenta, M., Lukauskas, S., Marscha, S., Leuchtenberger, S., Fuchs, H., Gaius-Durner, V., Hrabe de Angelis, M., Bultmann, S., Rando, O.J., Guccione, E., Kellner, S.M. and Schneider, R. (2020). METTL6 is a tRNA m3C methyltransferase that regulates pluripotency and tumor cell growth. Science Advances, 6, eaaz4551
- Zoch, A., Auchynnikava, T., Berrens, R. V., Kabayama, Y., Schöpp, T., Heep, M., Vasiliauskaitė, L., Pérez-Rico, Y. A., Cook, A. G., Shkumatava, A., Rappsilber, J., Allshire, R. C. and O'Carroll, D. (2020). SPOCD1 is an essential executor of piRNA-directed 1 de novo DNA methylation. Nature 584, 635-639.
- Ignatova, V.V., Stolz, P., Kaiser, S., Gustafsson, T.H., Rico Lastres, P., Sanz-Moreno, A., Cho, Y.L., Amarie, O.V., Aguilar-Pimentel, A., Klein-Rodewald, T., Calzada-Wack, J., Becker, L., Marschall, S., Kraiger, M., Garrett, L., Seisenberger, C., Hölter, S.M., Borland, K., Van De Logt, E., Jansen, P., Baltissen, M.P., Vermeulen, M., Wurst, W., Gailus-Durner, V., Fuchs, H., Hrabe de Angelis, M., Rando, O.J., Kellner, S.M., Bultmann, S. and Schneider. R. (2020). The rRNA m6A methyltransferase METTL5 regulates pluripotency and developmental programmes. Genes and Development , 34, 715-729.
- Morgan, M., Much, C., DiGiacomo, M., Azzi, C., Ivanova, I., Vitsios, D. M., Pistolic, J., Collier, P., Ventura De Oliveira Moreira, P., Benes, V., Enright, A. J. and O'Carroll, D. (2017). mRNA 3ʹ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome Nature 548, 347-351.
