Project description
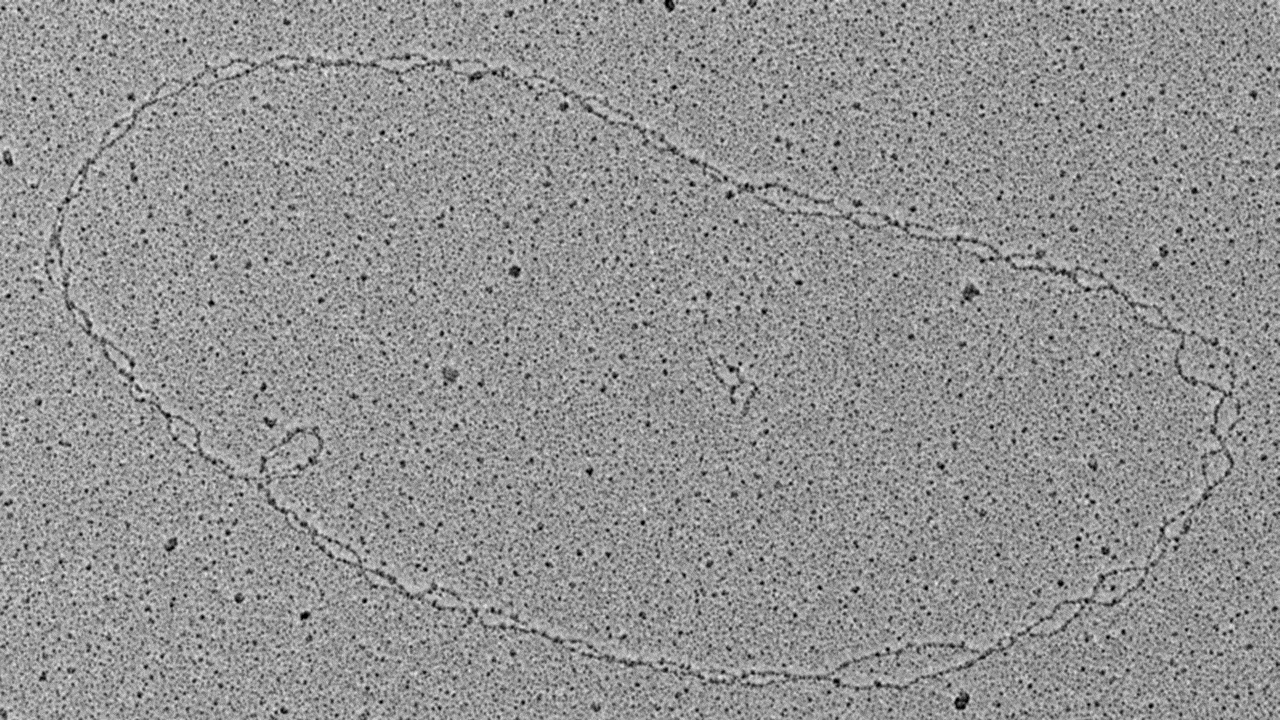
DNA replication is a fundamental process initiated at specialized regions named origins of replication. In yeast Saccharomyces cerevisiae origins, short (~200bp) autonomous replicating sequence (ARS) are present containing a conserved 11-bp AT-rich sequence named ARS consensus sequence (ACS), which serves as the binding site of the origin recognition complex (ORC). Although ARS sequences are conserved, they display striking differences in their intrinsic replication properties regarding timing and firing efficiency. Similar to all other eukaryotes, yeast replication origins can thus be classified into two main categories: early-efficient and late-inefficient origins. However, the molecular basis for these intrinsic differences remains largely elusive. Interestingly, the ARS confer a nucleosome-free region (NFR) flanked by well-positioned nucleosomes at precise intervals on both sides of the origin. As part of this project, we will purify defined chromatin domains of selected early-efficient or late-inefficient origins using an established locus-specific chromatin isolation protocol. We will then determine the functional relevance of this defined origin-proximal nucleosome positioning and capture the heterogeneity of nucleosome configurations by electron microscopy and DNA methylation footprinting using Nanopore-sequencing. These complementary state-of-the art single molecule datasets will not only address the question if the frequency of certain nucleosome configurations correlate with the experimentally observed differences of early-firing efficient versus late-firing inefficient origins, but also further our understanding of the molecular basis of the replication timing program in eukaryotic genomes.
Relevant literature
- Saldivar, J.C., Hamperl, S., Bocek, M.J., Chung, M., Bass, T.E., Cisneros-Soberanis, F., Samejima, K., Xie, L., Paulson, J.R., Earnshaw, W.C., Cortez, D., Meyer, T. and Cimprich, K.A. (2018). An intrinsic S/G2 checkpoint enforced by ATR. Science doi: 10.1126/science.aap9346.
- Hamperl, S., Bocek M., Saldivar, J. C., Swigut T., and Cimprich, K. A. (2017). Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 170, 774–786.
- Brown, C.R., Eskin, J.A., Hamperl, S., Griesenbeck, J., Jurica, M.S., and Boeger, H. (2015). Chromatin structure analysis of single gene molecules by psoralen cross-linking and electron microscopy. Methods Mol. Biol. 1228, 93–121.
- Hamperl, S., Brown, C.R., Perez-Fernandez, J., Huber, K., Wittner, M., Babl, V., Stöckl, U., Boeger, H., Tschochner, H., Milkereit, P., et al. (2014a). Purification of specific chromatin domains from single-copy gene loci in Saccharomyces cerevisiae. Methods Mol. Biol. 1094, 329–341.
- Hamperl, S., Brown, C.R., Garea, A.V., Perez-Fernandez, J., Bruckmann, A., Huber, K., Wittner, M., Babl, V., Stoeckl, U., Deutzmann, R., et al. (2014b). Compositional and structural analysis of selected chromosomal domains from Saccharomyces cerevisiae. Nucleic Acids Res. 42, e2.
