Project description
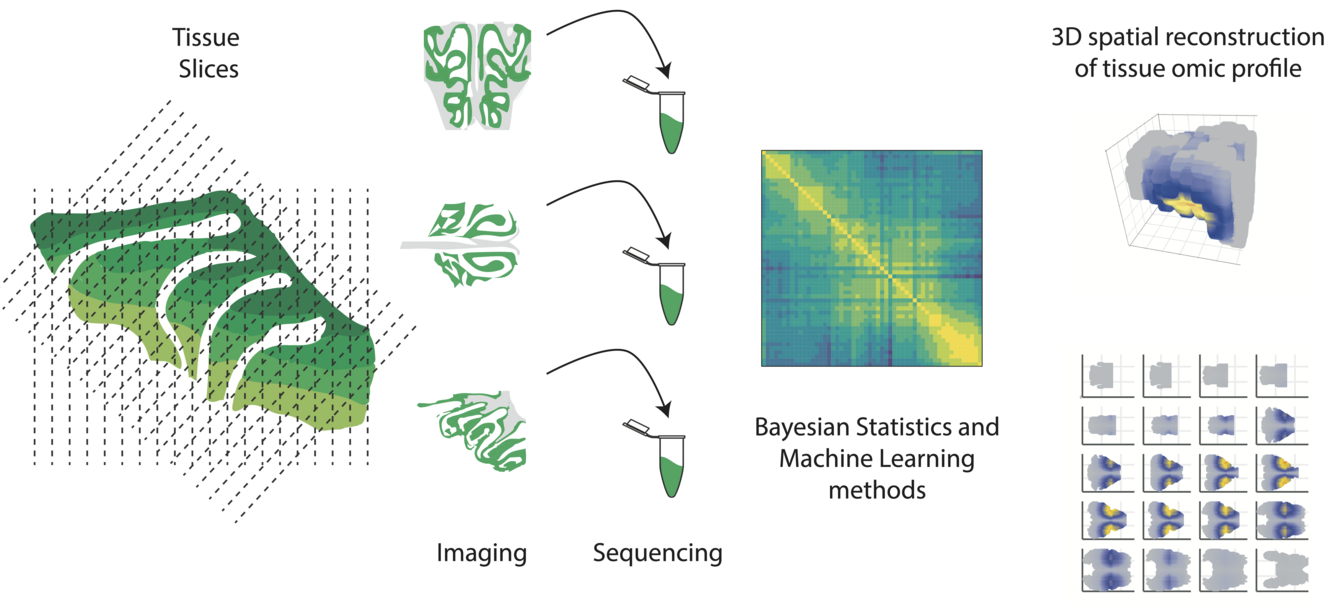
Omics data have opened new frontiers in the molecular description and understanding of how cells work. Nowadays, it is possible to probe gene activity, DNA accessibility and other molecular phenotypes with a single-cell resolution. However, spatial information, which is key to understanding how cells form organs and tissues, is typically lost. The molecular profiling of cells in their spatial context is the focus of spatial omics techniques, which are being developed at a rapid pace and have multiple applications in biomedicine and fundamental biology1–3. One of such techniques consists in analyzing uni-dimensional slices taken across different axes of a tissue. Computational methods are subsequently required to reconstruct the molecular profiles in the tri-dimensional space, similar to how tomography imaging works4,5.
In principle, spatial omics techniques are very powerful, but robust and statistically sound computational models are required to fully exploit their potential. Bayesian models are particularly suitable for this task, given their ability to take into account prior information on the geometry of the tissue and to provide a quantification of the uncertainty.
This project has two main aims: (i) develop new methods from Bayesian statistics to analyze spatial omic datasets based on tissue slicing; (ii) apply these techniques to study fundamental biological questions using existing and newly collected data from a variety of tissues.
During the method development step several questions will be addressed, for instance: how can we incorporate prior information on the geometry and cell composition of the tissue? Can we use the model to optimize experimental design, e.g. to estimate how many slices should be collected and in which orientations? How can we compare data collected across different biological conditions? What is the best way to identify "zones" of tissues sharing similar properties? How can we exploit single-cell data, when available? What adjustments do we need in order to make the model work with different types of omic data (e.g., transcriptomic, epigenomic)? All the methods generated will be implemented as open- source software tools.
We will collaborate closely with several experimental labs and apply our computational techniques to datasets collected from mouse olfactory epithelium, brain and bone. In addition to providing testing datasets for our methods, these collaborations will represent a unique opportunity to explore fundamental biological questions ranging from the functioning of the olfactory system, to the response of brain tissue to injury and the spatial architecture of bone tissue.
Relevant literature
- Saviano, A., Henderson, N. C. & Baumert, T. F., (2020). Single-cell genomics and spatial transcriptomics: Discovery of novel cell states and cellular interactions in liver physiology and disease biology. J. Hepatol., 73(5):1219-1230, doi:10.1016/j.jhep.2020.06.004.
- Moncada, R., Barkley D., Wagner F., Chiodin M., Devlin J. C., Baron M., Hajdu C. H., Simeone D. M., Yanai I. (2020). Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissu architecture in pancreatic ductal adenocarcinomas. Nature Biotechnology, 38, 333–342
- Crosetto, N., Bienko, M. & van Oudenaarden, A., (2015). Spatially resolved transcriptomics and beyond. Nat. Rev. Genet., 16, 57–66
- Junker, J. P., Noël E. S., Guryev V., Peterson K. A., Shah G., Huisken J., McMahon A. P., Berezikov E., Bakkers J., Van Oudenaarden A. (2014). Genome-wide RNA Tomography in the Zebrafish Embryo. Cell, 159, 662– 675
- Schede, H. H., Schneider C. G., Stergiadou J., Borm L. E., Ranjak A., Yamawaki T. M., David F. P. A., Lönnerberg P., Laurent G., Tosches M A., Codeluppi S., La Manno G. (2020). Spatial tissue profiling by imaging-free molecular tomography. bioRxiv, 2020.08.04.235655
- Vallejos, C. A., Risso D., Scialdone A., Dudoit S., Marioni J. (2017) Normalizing single-cell RNA sequencing data: Challenges and opportunities. Nat. Methods 14, 565-571
