Project description
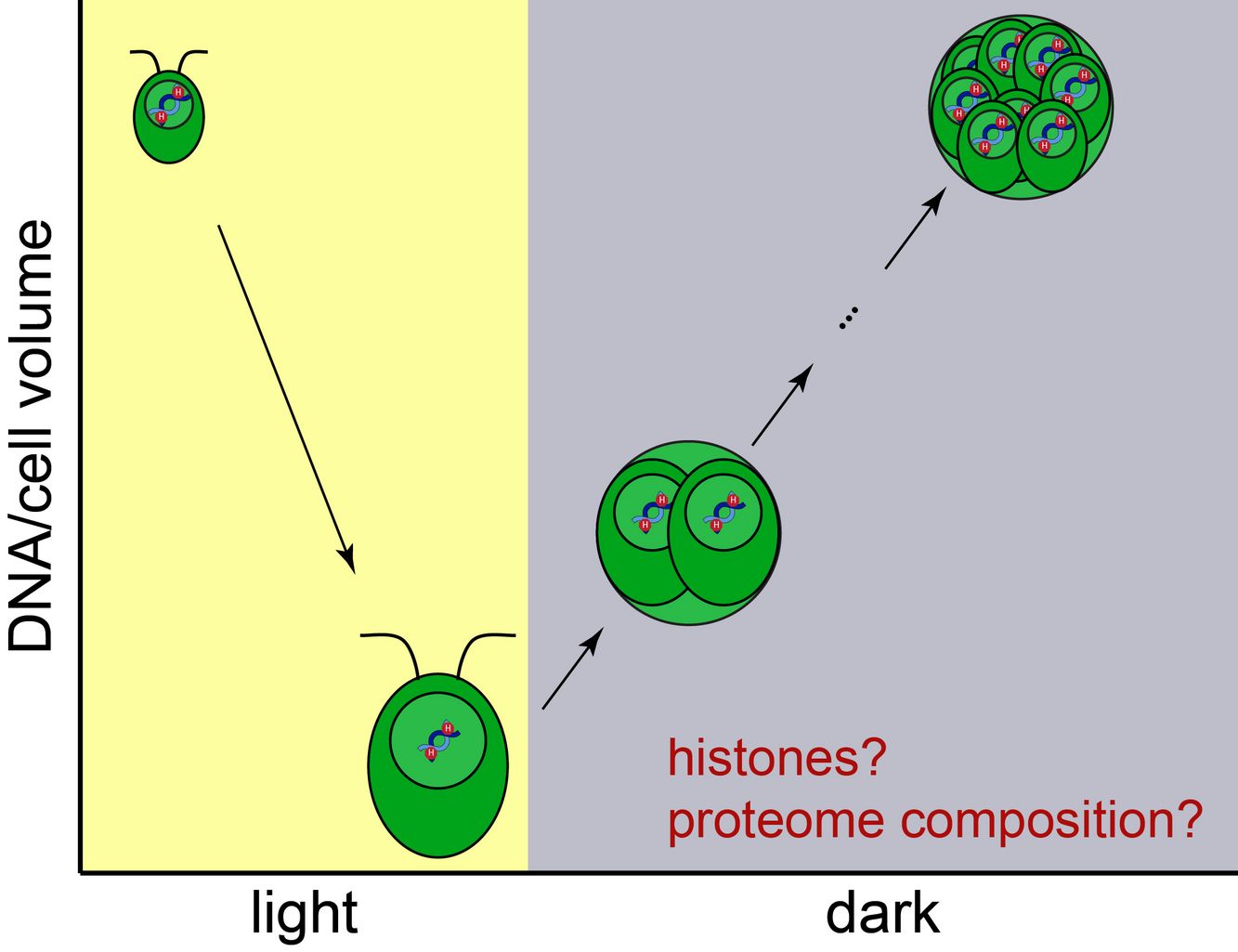
Size is a fundamental cellular property that is not only tightly linked to cell function and growth, but also sets the scale of all intracellular processes. Studying cell-size dependent regulation of chromatin-related processes in traditional model systems such as yeast or cell culture is complicated by the fact that cells tightly control their own size, limiting the variation of cell volume within a population of cells to about a factor of two. By contrast, the green alga Chlamydomonas reinhardtii exhibits a diurnal growth pattern, where during light hours cells can grow more than 30 fold without dividing, and then undergo a series of cleavage divisions during dark hours. These dramatic changes in cell volume – which can be tuned by modifying growth conditions – make Chlamydomonas the ideal model organism to study fundamental cell-size-dependent regulation.
Aim of this project is to establish Chlamydomonas reinhardtii as a model system to study cell-size-dependent processes, in particular the regulation of chromatin with cell size. Each of the multiple cleavage divisions results in a reduction of cellular and nuclear volume by a factor of two. At the same time, DNA concentration doubles with each division, which likely requires an exponential increase of the number of chromatin proteins, including histones. However, how cells would achieve such a cell-size-dependent adaptation of the proteome, and how the dramatic changes in nuclear volume affect chromatin organization and epigenetic regulation, and as a consequence transcription, is unknown. To address these questions, we will apply a combination of cell biological experiments and quantitative fluorescence microscopy, including single molecule FISH. Using Chlamydomonas as a model, we aim to reveal fundamental regulatory principles governing cell-size-dependent regulation across species.
Relevant literature
- Claude K-L, Bureik D, Adarska P, Singh A, Schmoller KM, 2020, Transcription coordinates histone amounts and genome content, bioRxiv, doi.org/10.1101/2020.08.28.272492
- Kukhtevich IV, Lohrberg N, Padovani F, Schneider R, Schmoller KM, 2020, Cell size sets the diameter of the budding yeast contractile ring, Nature Communications, 22, 2952
- Li Y, Liu D, López-Paz C, Olson BJSC, Umen JG, 2016, A new class of cyclin dependent kinase in Chlamydomonas is required for coupling cell size to cell division, eLIFE, 5, e10767
- Schmoller KM, Skotheim JM, 2015, The biosynthetic basis of cell size control, Trends in Cell Biology, 25, 12, 793-802
- Schmoller KM, Turner JJ, Kõivomägi M, Skotheim JM, 2015, Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size, Nature, 526, 268-272
