Project description
Cells must constantly balance and adjust the precise composition of their proteome in response to
external and internal cues. For many proteins, this requires matching their synthesis rates with the
size and growth rate of the cell. For example, ribosomes and other biosynthetic machinery are
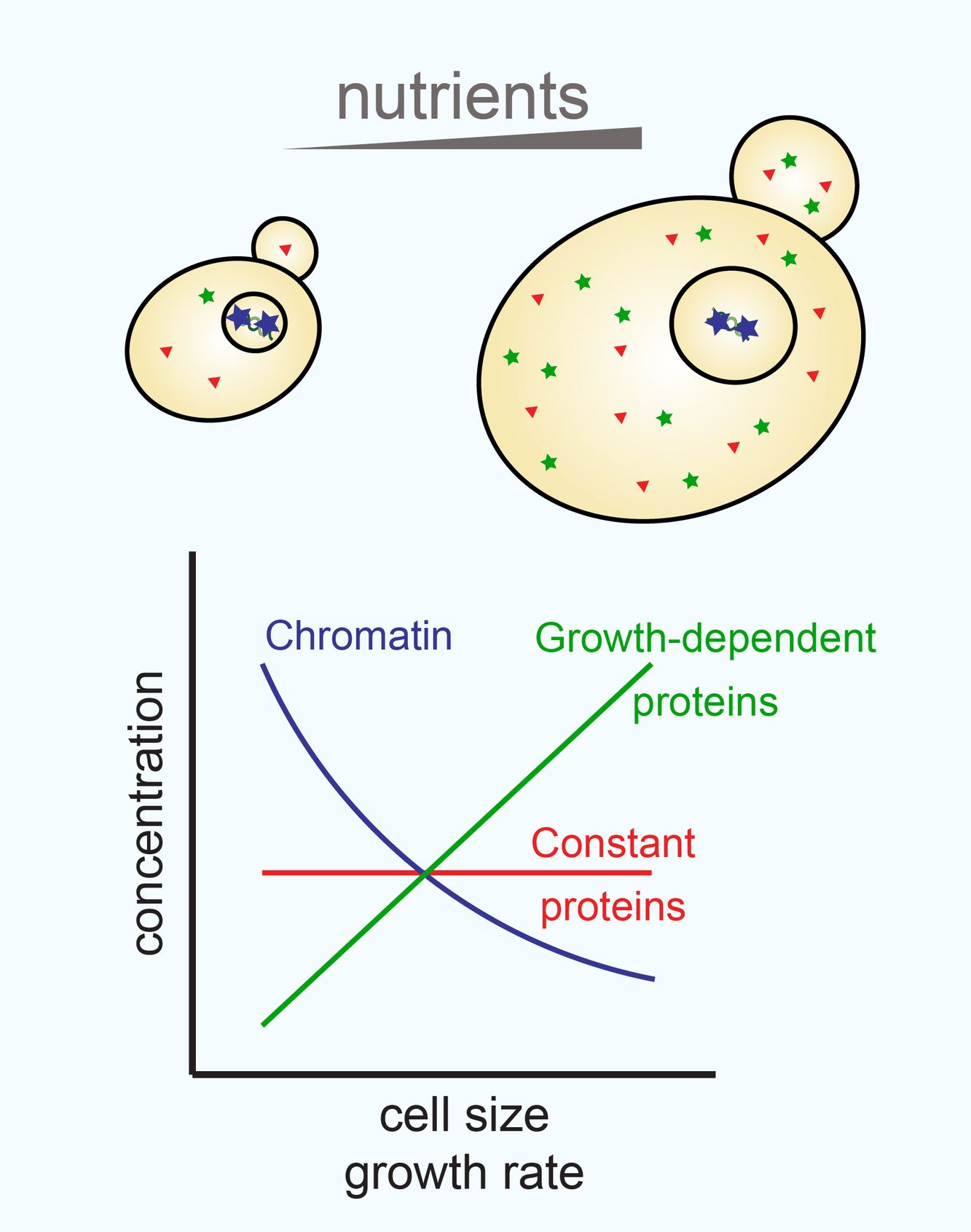
tightly correlated with growth across a range of conditions. However, specific subsets of proteins
need to be regulated differently – one of the best examples of this is chromatin. Chromatinassociated
proteins, including core histone proteins, are directly coupled to DNA amount, and
therefore cell cycle, but are independent of cell size or growth rate. This ensures that histones are
kept at a 1-to-1 ratio with the genome, such that the structure, composition, and function of
chromatin can always be maintained.
Due to this variable requirement for different protein sectors, the proteome is constantly
remodelled whenever cell size, cellular growth rate or cell cycle change. Ensuring correct proteome
composition is therefore particularly challenging when moving between different environmental
conditions, because, depending on the nutrients, all three of these parameters will change.
Our preliminary results show that in budding yeast, maintaining constant amounts of histones
across different environmental conditions requires both transcriptional and post-transcriptional
regulatory controls. In this project, we will use a combination of single-cell imaging, quantitative
functional genomics and high-throughput proteomics to determine how the production of other key
chromatin components is regulated as cells move between different nutritional states. In addition,
we will also examine more generally how other proteome sectors are regulated in response to
altered environmental conditions. After characterizing the proteome composition and revealing
whether the responsible regulation is transcriptional or post-transcriptional genome-wide, we will
focus on specific genes with distinct regulation to identify the underlying molecular mechanisms.
Adaptation to changing environments is a ubiquitous challenge for all eukaryotes. This project will
uncover the underlying regulatory principles by exploiting the powerful molecular tools available for
the model eukaryote budding yeast, including quantitative genomics approaches and multigenerational
live-cell imaging.
Relevant literature
Swaffer MP, Marinov GK, Zheng H, Jones AW, Greenwood J, Kundaje A, Snijders AP, Greenleaf
WJ, Reyes-Lamothe R, Skotheim JM, 2022, RNA polymerase II dynamics and mRNA stability
feedback determine mRNA scaling with cell size, bioRxiv,
https://doi.org/10.1101/2021.09.20.461005
Seel A, Padovani F, Finster A, Mayer M, Bureik D, Osman C, Klecker T, Schmoller KM, 2021,
Regulation with cell size ensures mitochondrial DNA homeostasis during cell growth, bioRxiv,
doi.org/10.1101/2021.12.03.471050
Swaffer MP, Kim J, Chandler-Brown D, Langhinrichs M, Marinov GK, Greenleaf WJ, Kundaje A,
Schmoller KM, Skotheim JM, 2021, Transcriptional chromatin-based partitioning mechanisms
uncouple protein scaling from cell cell size, Molecular Cell, 81, 1-15
Claude K-L, Bureik D, Chatzitheodoridou D, Adarska P, Singh A, Schmoller KM, 2020,
Transcription coordinates histone amounts and genome content, Nature Communications, 12,
4202
Kukhtevich IV, Lohrberg N, Padovani F, Schneider R, Schmoller KM, 2020, Cell size sets the
diameter of the budding yeast contractile ring, Nature Communications, 22, 2952
